Splicing and genetic susceptibility to cancer
Lab B-8Head: Eladio A. Velasco-Sampedro (Research Scientist, CSIC), ORCID , Scopus
Lab members:
Elena Bueno-Martínez (Assistant Professor, Universidad de Valladolid)Alicia García Álvarez (Technician, CSIC)
Lara Sanoguera Miralles (Predoctoral student, Asociación Española Contra el Cáncer, 2019-2023)
Inés Llinares Burguet (Predoctoral Fellowship, Consejería de Educación, Junta de Castilla y León, 2022-2025)
Line of Research
Our interest is focused on Hereditary Breast and Ovarian Cancer (HBOC) that is characterized by a high genetic heterogeneity. Loss-of-function variants in at least 8 genes (BRCA1, BRCA2, PALB2, BARD1, RAD51C, RAD51D, ATM, and CHEK2) have been recently reported with a significant association with breast cancer risk.
Most of the cancer susceptibility genes are involved in the DNA repair pathway in order to keep the genomic integrity. The two principal genes, BRCA1 and BRCA2, account for only 16% of familial breast cancer risk, whereas the rest of the genes and SNPs (GWAS studies) contribute up to 50% of the familial risk. Genetic testing of BRCA1 and BRCA2 provides essential information for the clinical management of HBOC families since it allows the detection of asymptomatic mutation carriers and facilitates preventive decision-making.
On the other hand, ~20% of patients carry a DNA variant of unknown clinical significance (VUS) since it is not known whether they are neutral or disease-causing, hampering genetic diagnostic and, therefore, disease prevention. Pathogenic mutations are often predicted on the basis of their impact on protein function but other gene expression steps, such as transcription and splicing, may be disrupted by DNA variants and involved in a disease.
Splicing is a central process of gene expression whereby introns are excised and exons are joined sequentially. Splicing in YouTube: https://www.youtube.com/watch?v=FVuAwBGw_pQ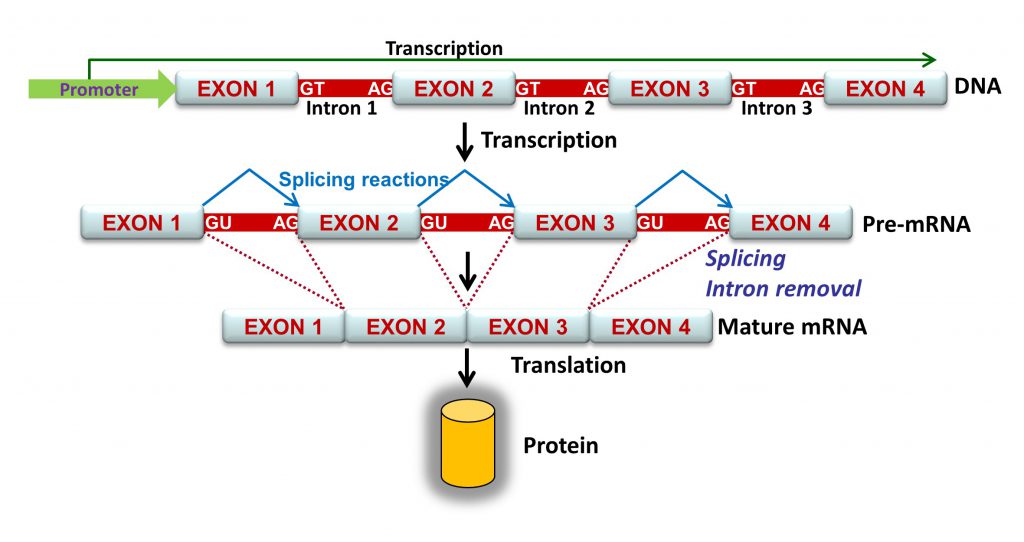
Intron removal is guided by a large number of splicing factors, ribonucleoproteins and a complex array of sequences including acceptor and donor sites, polypyrimidine tract and branch point, as well as exonic and intronic splicing enhancers and silencers that stimulate or repress exon inclusion into the mature mRNA, respectively. Consequently, all these elements are potential targets of spliceogenic variants.
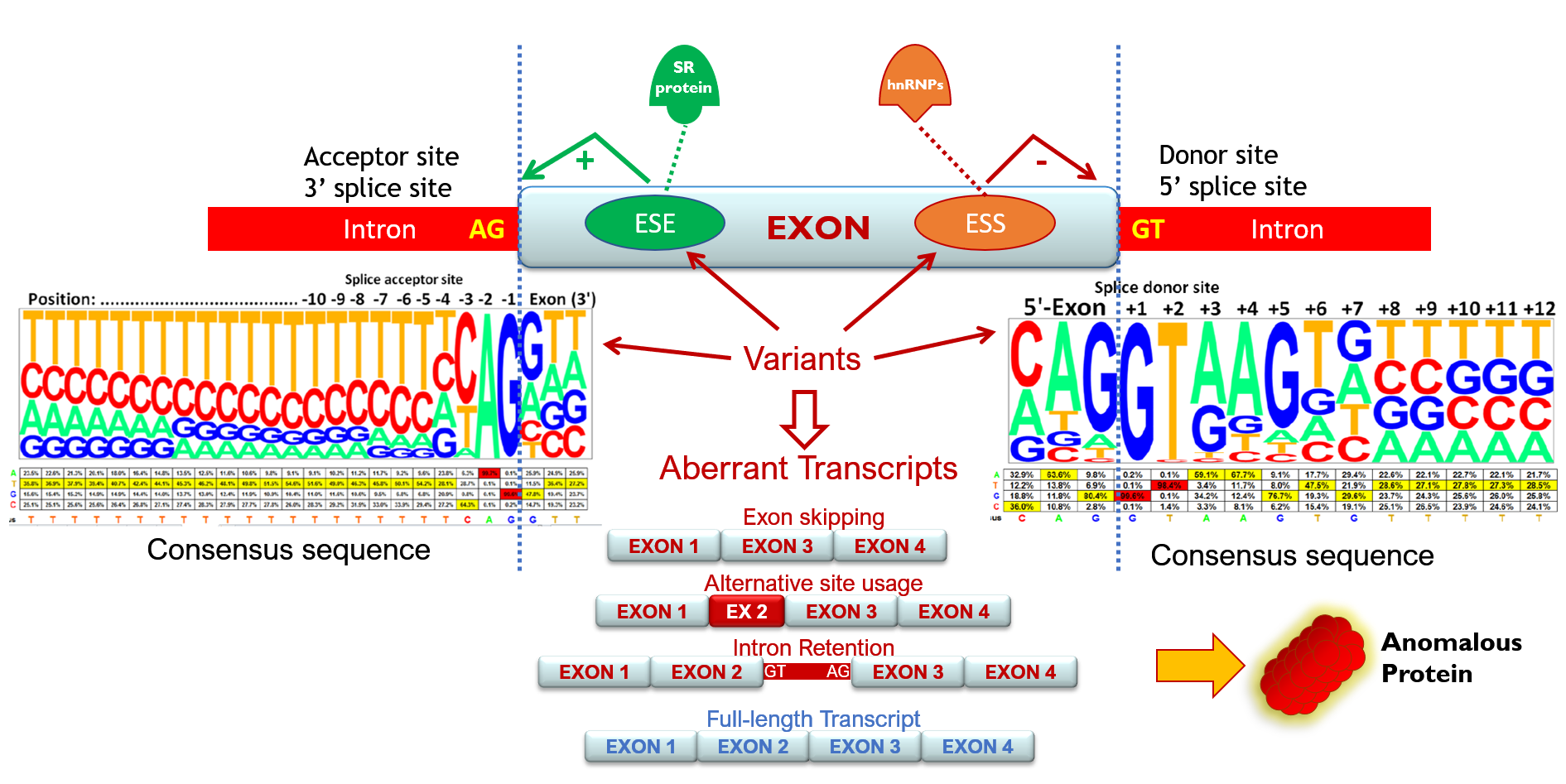
These alterations remain often hidden as patient RNA assays to characterize them are not available. Reclassification of a DNA variant as deleterious increases the number of patients and families who may benefit from tailored prevention protocols. Consequently, the identification of pathogenic variants will contribute to breast cancer prevention, therapy, and surveillance as well as to a better knowledge of the breast cancer genetics that will even allow the development of new alternative therapies.
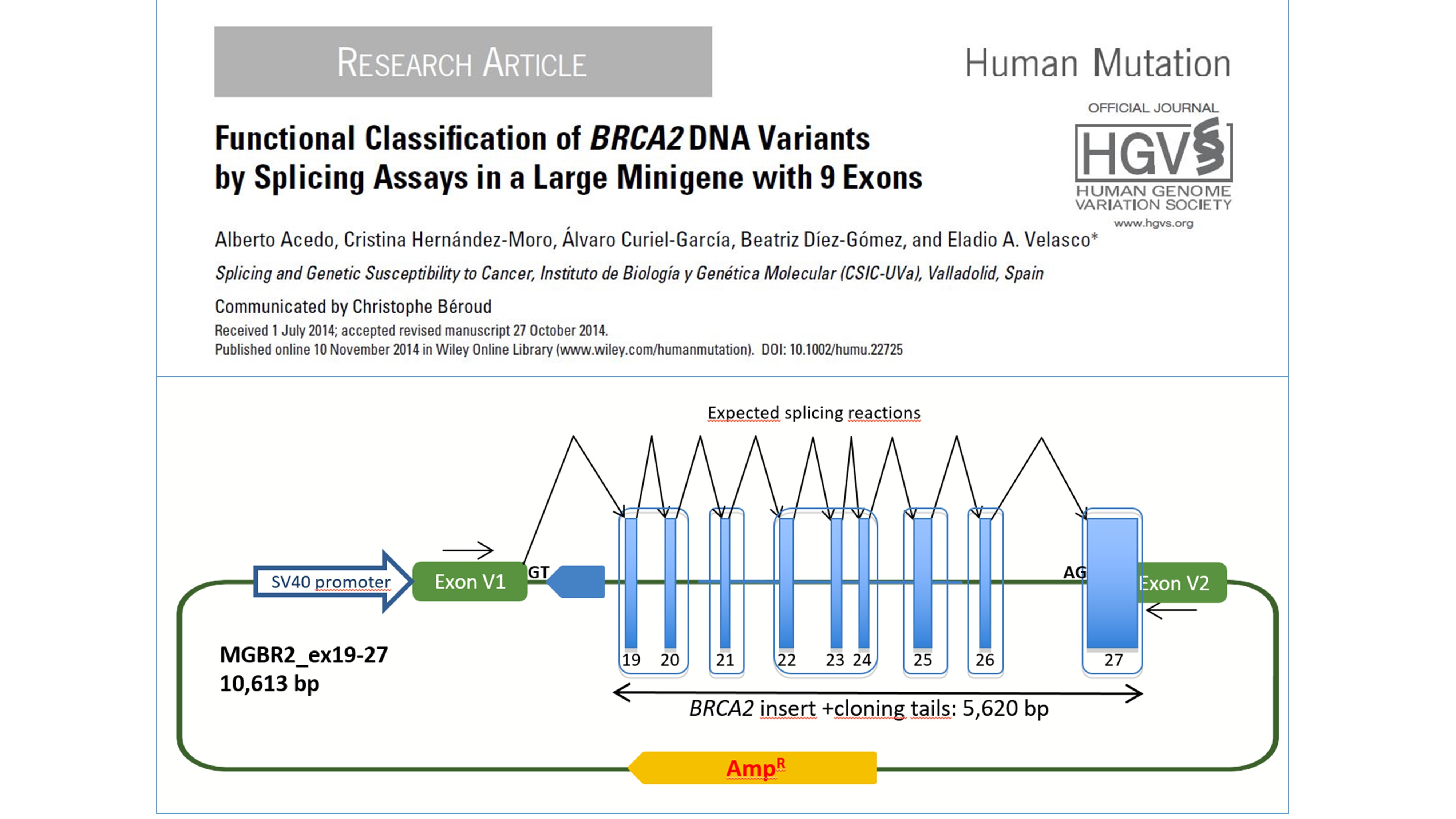
Our main line of research aims to investigate the role of mis-splicing of breast cancer genes in HBOC. Hybrid minigenes in splicing reporter plasmid are our main tool of the laboratory. They constitute a useful technology for both, the basic knowledge of splicing regulatory mechanisms and the study of the correlation between mutation, defective splicing and disease. In fact, we have developed and patented a new splicing vector, pSAD (Splicing And Disease; patent #P201231427: “Plasmid pSAD for splicing functional assays”), where there have been constructed a complete set of Breast Cancer minigenes, including BRCA2, BRCA1, CHEK2, PALB2, ATM, RAD51C, RAD51D and BARD1. Therefore, we can potentially assay any candidate variant in these genes to study its impact on splicing. Actually, we have already analyzed ~1,000 variants by minigene assays. The identification of abnormal splicing patterns in the susceptibility genes will contribute to elucidate the genetic susceptibility spectrum of breast/ovarian cancer.
The largest minigene ever reported with BRCA2 exons 19 to 27.
We have also set up a splicing-minigene facility to study any human gene under the splicing viewpoint. In fact, we have already developed minigenes of MLH1 (Lynch syndrome), SERPINA1 (Severe Alpha-1 Antitrypsin Deficiency), COL1A1 (Osteogenesis Imperfecta), CHD7 (Intellectual Disability), GRN (Frontotemporal Dementia) and UGT1A1 (Gilbert’s and Crigler-Najjar syndromes).
OBJECTIVES
The main objective of our line of research is to clarify the genetic susceptibility landscape of HBOC. As a translational research laboratory we aim that our findings have an impact on patient’s health by improving the molecular diagnostics and prevention of this disease.
Specific goals:
Correlation between aberrant splicing of breast cancer susceptibility genes and hereditary breast/ovarian cancer.
Pre-mRNA processing (splicing):-
- Study of the correlation between genetic variants, aberrant splicing patterns of breast cancer genes and HBOC: splice-site variants, use of cryptic sites and generation of de novo sites.
- Regulation of long exons of breast cancer genes and implications in HBOC.
- Regulation of non-canonical splice sites: GC donor sites in DNA repair genes such as BRCA2, FANCA and PALB2. Splicing impact of +2T>C variants in exons of susceptibility genes.
Financial Support
- Instituto de Salud Carlos III. Ministerio de Ciencia, Innovación y Universidades. Proyecto de Investigación en Salud, Referencia: PI23/00047 (2024-2026). Desregulación del splicing de macroexones y microexones en genes de susceptibilidad a cáncer de mama. Investigador principal: Eladio A. Velasco Sampedro. Total amount: 91.250 €.
- Instituto de Salud Carlos III. Ministerio de Ciencia e Innovación. Proyecto de Investigación en Salud, Referencia: PI20/00225 (2021-2023). "Splicing regulation of alternative and atypical exons of breast cancer genes in physiological and DNA damage conditions: implications in disease susceptibility" . Investigador principal: Eladio A. Velasco Sampedro. Total amount: 93.170 €.

- Consejería de Educación, Junta de Castilla y León. "Desregulación del splicing en cáncer de mama hereditario: análisis funcional de genes de susceptibilidad mediante minigenes híbridos". Investigador principal: Eladio A. Velasco Sampedro. Total amount: 120.000 €. REF. CSI242P18. Actuación cofinanciada P.O. FEDER 2014-2020



- Instituto de Salud Carlos III. Ministerio de Economía y Competitividad. Proyecto de Investigación en Salud, Referencia: PI17/00227 (2018-2020). "Aberrant splicing in hereditary breast cancer: Functional analysis of susceptibility genes by hybrid minigenes" . Investigador principal: Eladio A. Velasco Sampedro. Total amount: 99.220 €.

- European Commision, Project ID 634935 (2015-2020): "Breast Cancer Risk after Diagnostic Gene Sequencing (BRIDGES)". Partner #18, Investigador principal: Eladio A. Velasco Sampedro. Total amount: 75.000 €.

- Instituto de Salud Carlos III. Ministerio de Economía y Competitividad. Proyecto de Investigación en Salud, Project ID PI13/01749 (2014-2016): "Alterations of transcription and splicing of the BRCA1 and BRCA2 genes and genetic susceptibility to Breast and Ovarian Cancer". Total amount: 74.838,5 €.

- Instituto de Salud Carlos III. Ministerio de Ciencia e Innovación. Proyecto de Investigación en Salud PI10/02910 (2011-2013): “Análisis de secuencias reguladoras de splicing (enhancer y silenciadores) de BRCA1 y BRCA2 mediante minigenes híbridos: splicing aberrante y cáncer de mama/ovario. Análisis global de los patrones de splicing en pacientes BRCA negativos.”

- Consejería de Educación, Junta de Castilla y León (Project ID CSI090U14): "Impact of transcriptional and splicing regulatory mutations of tumor-suppressor genes on molecular diagnostics and prevention of breast cancer"

- Asociación Española Contra el Cáncer (Valladolid)

Publications
-
- Llinares-Burguet I, Sanoguera-Miralles L, Valenzuela-Palomo A, García-Álvarez A, Bueno-Martínez E, Velasco-Sampedro EA. Splicing Dysregulation of Non-Canonical GC-5' Splice Sites of Breast Cancer Susceptibility Genes ATM and PALB2. Cancers (Basel). 2024;16:3562. doi: 10.3390/cancers16213562. PMID: 39518003; PMCID: PMC11545216.
- Carmona-Carmona CA, Zini P, Velasco-Sampedro EA, Cózar-Castellano I, Perdomo G, Caloca MJ. β2-Chimaerin, a GTPase-Activating Protein for Rac1, Is a Novel Regulator of Hepatic Insulin Signaling and Glucose Metabolism. Molecules. 2024;29:5301. doi: 10.3390/molecules29225301. PMID: 39598690; PMCID: PMC11597029.
- Sanoguera-Miralles L, Llinares-Burguet I, Bueno-Martínez E, Ramadane-Morchadi L, Stuani C, Valenzuela-Palomo A, García-Álvarez A, Pérez-Segura P, Buratti E, de la Hoya M, Velasco-Sampedro EA. Comprehensive splicing analysis of the alternatively spliced CHEK2 exons 8 and 10 reveals three enhancer/silencer-rich regions and 38 spliceogenic variants. J Pathol. 2024;262:395-409. doi: 10.1002/path.6243. PMID: 38332730.
- Sanoguera-Miralles L, Valenzuela-Palomo A, Bueno-Martínez E, Esteban-Sánchez A, Lorca V, Llinares-Burguet I, García-Álvarez A, Pérez-Segura P, Infante M, Easton DF, Devilee P, Vreeswijk MPG, de la Hoya M, Velasco-Sampedro EA. Systematic Minigene-Based Splicing Analysis and Tentative Clinical Classification of 52 CHEK2 Splice-Site Variants. Clin Chem. 2024; 70:319-338. doi: 10.1093/clinchem/hvad125. PMID: 37725924.
- Valenzuela-Palomo A, Sanoguera-Miralles L, Bueno-Martínez E, Esteban-Sánchez A, Llinares-Burguet I, García-Álvarez A, Pérez-Segura P, Gómez-Barrero S, de la Hoya M, Velasco-Sampedro EA. Splicing Analysis of 16 PALB2 ClinVar Variants by Minigene Assays: Identification of Six Likely Pathogenic Variants. Cancers (Basel). 2022;14:4541. doi: 10.3390/cancers14184541. PMID: 36139699.
- Sanoguera-Miralles L, Bueno-Martínez E, Valenzuela-Palomo A, Esteban-Sánchez A, Llinares-Burguet I, Pérez-Segura P, García-Álvarez A, de la Hoya M, Velasco-Sampedro EA. Minigene Splicing Assays Identify 20 Spliceogenic Variants of the Breast/Ovarian Cancer Susceptibility Gene RAD51C. Cancers (Basel)2022;14:2960. doi: 10.3390/cancers14122960. PMID: 35740625.
- Bueno-Martínez E, Sanoguera-Miralles L, Valenzuela-Palomo A, Esteban-Sánchez A, Lorca V, Llinares-Burguet I, Allen J, García-Álvarez A, Pérez-Segura P, Durán M, Easton DF, Devilee P, Vreeswijk MPG, de la Hoya M, Velasco-Sampedro EA. Minigene-based splicing analysis and ACMG/AMP-based tentative classification of 56 ATM variants. J Pathol. 2022. doi:10.1002/path.5979. Online ahead of print. PMID: 35716007.
- Zhu L, Miao B, Dymerska D, Kuswik M, Bueno-Martínez E, Sanoguera-Miralles L, Velasco EA, Paramasivam N, Schlesner M, Kumar A, Yuan Y, Lubinski J, Bandapalli OR, Hemminki K, Försti A. Germline Variants of CYBA and TRPM4 Predispose to Familial Colorectal Cancer. Cancers (Basel) 2022;14:670. doi:10.3390/cancers14030670. PMID: 35158942.
- Valenzuela-Palomo A, Bueno-Martínez E, Sanoguera-Miralles L, Lorca V, Fraile-Bethencourt E, Esteban-Sánchez A, Gómez-Barrero S, Carvalho S, Allen J, García-Álvarez A, Pérez-Segura P, Dorling L, Easton DF, Devilee P, Vreeswijk MP, de la Hoya M, Velasco EA. Splicing predictions, minigene analyses, and ACMG-AMP clinical classification of 42 germline PALB2 splice-site variants. J Pathol. 2022;256:321-334. doi: 10.1002/path.5839. PMID: 34846068.
- Asselta R, Duga S, Velasco EA, Buratti E.Editorial: RNA Splicing and Backsplicing: Disease and Therapy. Front Genet. 2020 Dec 8;11:626835. doi: 10.3389/fgene.2020.626835. PMID: 33363577.
- Bueno-Martínez E, Sanoguera-Miralles L, Valenzuela-Palomo A, Lorca V, Gómez-Sanz A, Carvalho S, Allen J, Infante M, Pérez-Segura P, Lázaro C, Easton DF, Devilee P, Vreeswijk MPG, de la Hoya M, Velasco EA. RAD51D Aberrant Splicing in Breast Cancer: Identification of Splicing Regulatory Elements and Minigene-Based Evaluation of 53 DNA Variants. Cancers (Basel) 2021;13:2845. doi: 10.3390/cancers13112845. PMID: 34200360.
- Sanoguera-Miralles L.; Valenzuela-Palomo A.; Bueno-Martínez E.; Llovet P.; Díez-Gómez B.; Caloca M.J.; Pérez-Segura P.; Fraile-Bethencourt E.; Colmena M.; Carvalho S.; Allen J.; Easton D.F.; Devilee P.; Vreeswijk M.P.G.; de la Hoya M.; Velasco E.A. Comprehensive Functional Characterization and Clinical Interpretation of 20 Splice-Site Variants of the RAD51C Gene. Cancers (Basel) 2020, 12:3771. doi: 10.3390/cancers12123771. PMID: 33333735.
- Gailite L, Valenzuela-Palomo A, Sanoguera-Miralles L, Rots D, Kreile M and Velasco EA. UGT1A1 Variants c.864+5G>T and c.996+2_996+5del of a Crigler-Najjar Patient Induce Aberrant Splicing in Minigene Assays. Front. Genet. 2020; 11:169. doi: 10.3389/fgene.2020.00169. Front Genet 2020; 11: 169
- Casado-Medrano V, Barrio-Real L, Gutiérrez-Miranda L, González-Sarmiento R, Velasco EA, Kazanietz MG, Caloca MJ. Identification of a truncated β1-chimaerin variant that inactivates nuclear Rac1. J Biol Chem. 2020; 295:1300-1314. doi: 10.1074/jbc.RA119.008688
- Fraile-Bethencourt E, Valenzuela-Palomo A, Díez-Gómez B, Goina E, Acedo A, Buratti E, Velasco EA. Mis-splicing in breast cancer: identification of pathogenic BRCA2 variants by systematic minigene assays. J Pathol. 2019; 248: 409-420. doi: 10.1002/path.5268. [Pubmed]
- Fraile-Bethencourt E; Valenzuela-Palomo A; Díez-Gómez B; Caloca MJ; Gómez-Barrero S & Velasco EA. Minigene splicing assays identify 12 spliceogenic variants of BRCA2 exons 14 and 15. Frontiers in Genetics 2019; 10; // DOI: 10.3389/fgene.2019.00503.
- Lopez-Perolio I, Leman R, Behar R, Lattimore V, Pearson JF, Castéra L, Martins A, Vaur D, Goardon N, Davy G, Garre P, García-Barberán V, Llovet P, Pérez-Segura P, Díaz-Rubio E, Caldés T, Hruska KS, Hsuan V, Wu S, Pesaran T, Karam R, Vallon-Christersson J, Borg A, Investigators K, Valenzuela-Palomo A, Velasco EA, Southey M, Vreeswijk MPG, Devilee P, Kvist A, Spurdle AB, Walker LC, Krieger S, de la Hoya M. Alternative splicing and ACMG-AMP-2015-based classification of PALB2 genetic variants: an ENIGMA report. J Med Genet. 2019 DOI: jmedgenet-2018-105834.[Pubmed]
- Montalban G, Fraile-Bethencourt E, López-Perolio I, Pérez-Segura P, Infante M, Durán M, Alonso-Cerezo MC, López-Fernández A, Diez O, de la Hoya M, Velasco EA, Gutiérrez-Enríquez S (2018). Characterization of spliceogenic variants located in regions linked to high levels of alternative splicing: BRCA2 c.7976+5G > T as a case study. Human Mutation 2018 Jul 3. doi: 10.1002/humu.23583. Pubmed [Co-first and co-corresponding authors].
- Fraile-Bethencourt E, Valenzuela-Palomo A, Díez-Gómez B, Acedo A, Velasco EA (2018).Identification of 8 spliceogenic variants in BRCA2 exon 16 by minigene assays. Frontiers in Genetics 2018, 9: 188. DOI: 10.3389/fgene.2018.00188. https://www.frontiersin.org/articles/10.3389/fgene.2018.00188/full
- Fraile-Bethencourt E, Valenzuela-Palomo A, Díez-Gómez B, Infante M, Durán M, Marcos G, Lastra E, Gómez-Barrero S, Velasco EA (2018). Genetic dissection of the BRCA2 promoter and transcriptional impact of DNA variants. Breast Cancer Research and Treatment 2018. DOI: 10.1007/s10549-018-4826-7. https://www.ncbi.nlm.nih.gov/pubmed/29766361
- Fraile-Bethencourt E, Díez-Gómez B, Velásquez-Zapata V, Acedo A, Sanz DJ, Velasco EA (2017). Functional classification of DNA variants by hybrid minigenes: Identification of 30 spliceogenic variants of BRCA2 exons 17 and 18. PLoS Genetics 13(3):e1006691. doi: 10.1371/journal.pgen.1006691. http://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1006691
- Acedo A, Hernández-Moro C, Curiel-García Á, Díez-Gómez B, Velasco EA. Functional Classification of BRCA2 DNA Variants by Splicing Assays in a Large Minigene with 9 Exons. Human Mutation 2015; 36: 210–221. PMID: 25382762.
- Lara B, Martínez MT, Blanco I, Hernández-Moro C, Velasco EA, Ferrarotti I, Rodriguez-Frias F, Perez L, Vazquez I, Alonso J, Posada M, Martínez-Delgado B. Severe alpha-1 antitrypsin deficiency in composite heterozygotes inheriting a new splicing mutation QOMadrid. Respiratory Research 2014; 15: 125. PMID 25287719.
- Ruiz de Garibay G, Acedo A, García-Casado Z, Gutiérrez-Enríquez S, Tosar A, Romero A, Garre P, Llort G, Thomassen M, Díez O, Pérez-Segura P, Eduardo Díaz-Rubio E, Velasco EA, Caldés T, de la Hoya M. Capillary Electrophoresis Analysis of Conventional Splicing Assays: IARC Analytical and Clinical Classification of 31 BRCA2 Genetic Variants. Human Mutation 2014; 35: 53-57. PMID: 24123850
- Infante M, Duran M, Acedo A, Sánchez-Tapia EM, Díez-Gómez B, Barroso A, Garcia-Gonzalez M, Feliubadalo L, Lasa A, de la Hoya M; Esteban-Cardeñosa E; Díez O; Martínez-Bouzas C; Godino J; Teulé A; Osorio A; Lastra E; Gonzalez-Sarmiento R, Miner C, Velasco EA (2013). The highly prevalent BRCA2 mutation c.2808_2811del (3036delACAA) is located in a mutational hotspot and has multiple origins. Carcinogenesis 34: 2505-2511. PMID: 23929434
- Blanco A, de la HM, Osorio A, Diez O, Miramar MD, Infante M, Martinez-Bouzas C, Torres A, Lasa A, Llort G, Brunet J, Grana B, Perez SP, Garcia MJ, Gutierrez-Enriquez S, Carracedo A, Tejada MI, Velasco EA, Calvo MT, Balmana J, Benitez J, Caldes T, Vega A (2013) Analysis of PALB2 Gene in BRCA1/BRCA2 Negative Spanish Hereditary Breast/Ovarian Cancer Families with Pancreatic Cancer Cases. PLoS One 8:e67538.
- Perez-Cabornero L, Infante M, Velasco E, Lastra E, Miner C, Duran M (2013) Evaluating the Effect of Unclassified Variants Identified in MMR Genes Using Phenotypic Features, Bioinformatics Prediction, and RNA Assays. J Mol Diagn 15:380-390
- Acedo A, Sanz DJ, Duran M, Infante M, Perez-Cabornero L, Miner C, Velasco EA (2012) Comprehensive splicing functional analysis of DNA variants of the BRCA2 gene by hybrid minigenes. Breast Cancer Res 14: R87. PMID: 22632462.
- Perez-Cabornero L, Infante M, Velasco E, Lastra E, Acedo A, Miner C, Duran M (2011) Frequency of rearrangements in Lynch syndrome cases associated with MSH2: characterization of a new deletion involving both EPCAM and the 5' part of MSH2. Cancer Prev Res (Phila) 4:1556-1562
- Sanz DJ, Acedo A, Infante M, Duran M, Perez-Cabornero L, Esteban-Cardenosa E, Lastra E, Pagani F, Miner C, Velasco EA (2010). A high proportion of DNA variants of BRCA1 and BRCA2 is associated with aberrant splicing in breast/ovarian cancer patients. Clin Cancer Res 16:1957-1967. PMID: 20215541.
- Infante M, Duran M, Lasa A, Acedo A, de la HM, Esteban-Cardenosa E, Sanz DJ, Perez-Cabornero L, Lastra E, Miner C, Velasco EA (2010). Two founder BRCA2 mutations predispose to breast cancer in young women. Breast Cancer Res Treat 122:567-571
- Infante M, Duran M, Acedo A, Perez-Cabornero L, Sanz DJ, Garcia-Gonzalez M, Beristain E, Esteban-Cardenosa E, de la HM, Teule A, Vega A, Tejada MI, Lastra E, Miner C, Velasco EA (2010) BRCA1 5272-1G>A and BRCA2 5374delTATG are founder mutations of high relevance for genetic counselling in breast/ovarian cancer families of Spanish origin. Clin Genet 77:60-69
- Perez-Cabornero L, Velasco E, Infante M, Sanz D, Lastra E, Hernandez L, Miner C, Duran M (2009) A new strategy to screen MMR genes in Lynch Syndrome: HA-CAE, MLPA and RT-PCR. Eur J Cancer 45:1485-1493.
- Milne RL, Osorio A, Cajal TR, Vega A, Llort G, de la HM, Diez O, Alonso MC, Lazaro C, Blanco I, Sanchez-de-Abajo A, Caldes T, Blanco A, Grana B, Duran M, Velasco E, et al. (2008) The average cumulative risks of breast and ovarian cancer for carriers of mutations in BRCA1 and BRCA2 attending genetic counseling units in Spain. Clin Cancer Res 14:2861-2869
- Velasco E, Infante M, Duran M, Perez-Cabornero L, Sanz DJ, Esteban-Cardenosa E, Miner C (2007) Heteroduplex analysis by capillary array electrophoresis for rapid mutation detection in large multiexon genes. Nat Protoc 2:237-246