Servicio de splicing-minigenes
MINIGENE SERVICE
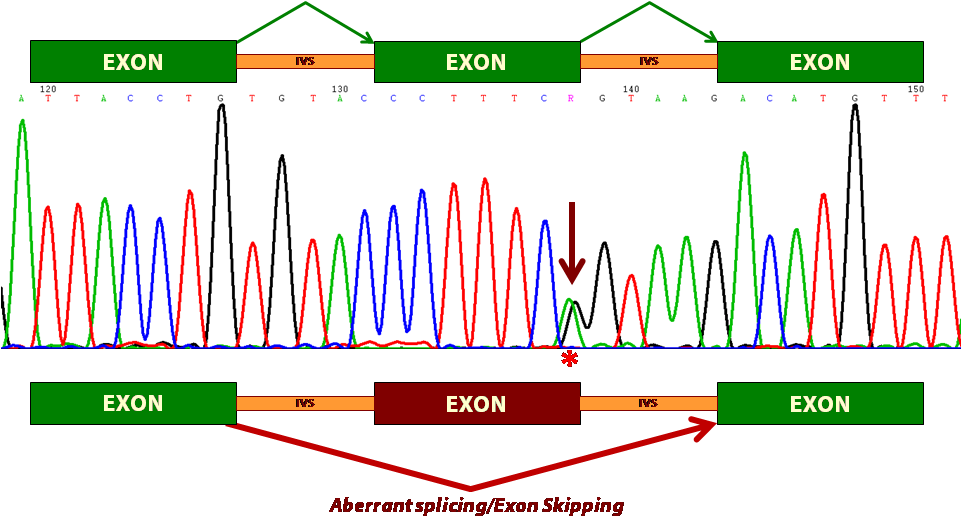
Impact of mutations on splicing. A DNA variant (indicated by an arrow in the sequence) of the donor site of BRCA2 exon 13 induces its loss (skipping) in the mature transcript. This causes an ORF change and the generation of a premature STOP codon.
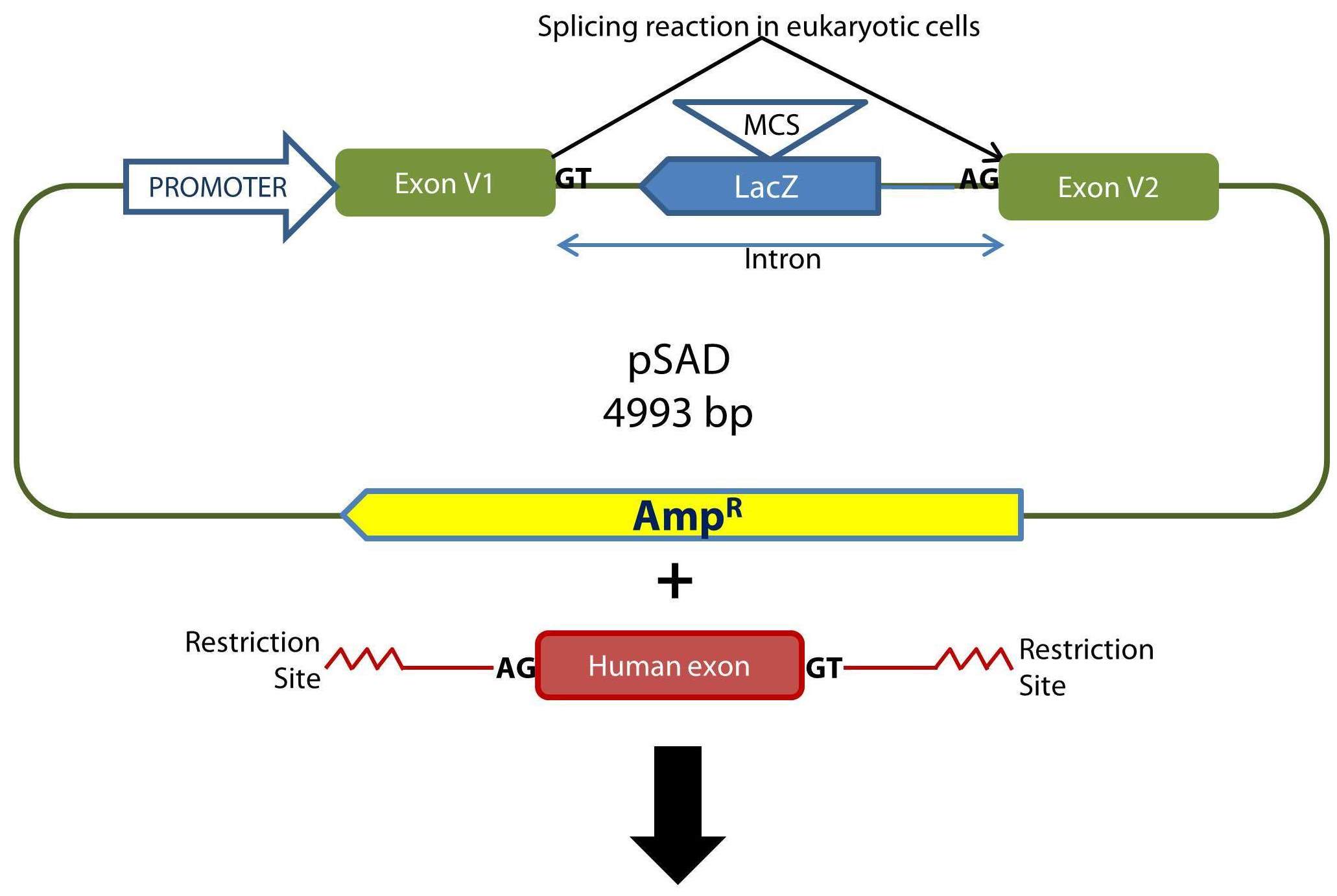
Functional analysis of DNA variants from disease-responsible genes can be carried out by direct patient RNA RT-PCR (usually lymphocyte RNA) or functional assay by hybrid minigenes. Patient RNA is not always available and often difficult to obtain so splicing reporter minigenes are useful alternative tools to study ex vivo the impact of a variant on splicing. The Splicing and Genetic susceptibility group of the IBGM (CSIC, PI Eladio A. Velasco) has developed a new splicing reporter plasmid (pSAD; https://docs.google.com/file/d/0B7Ne4lHDwcbubGpfdV9xX2NmQ3M/edit?usp=sharing) to carry out splicing functional assays by hybrid minigenes, which helps to identify variants with impact on splicing.
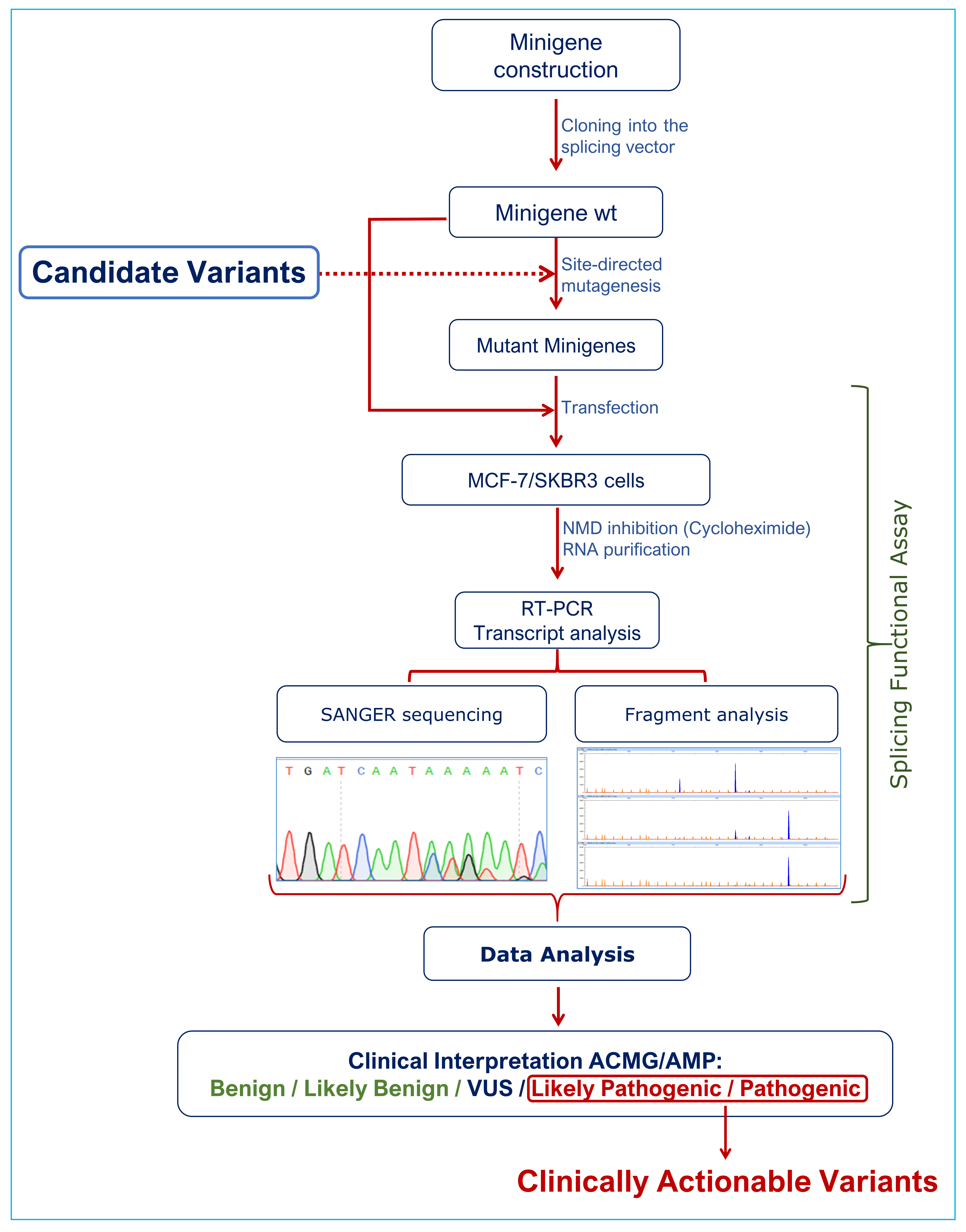
The protocol for a splicing functional analysis of a particular DNA variant of a disease-responsible gene is as simple as follows:
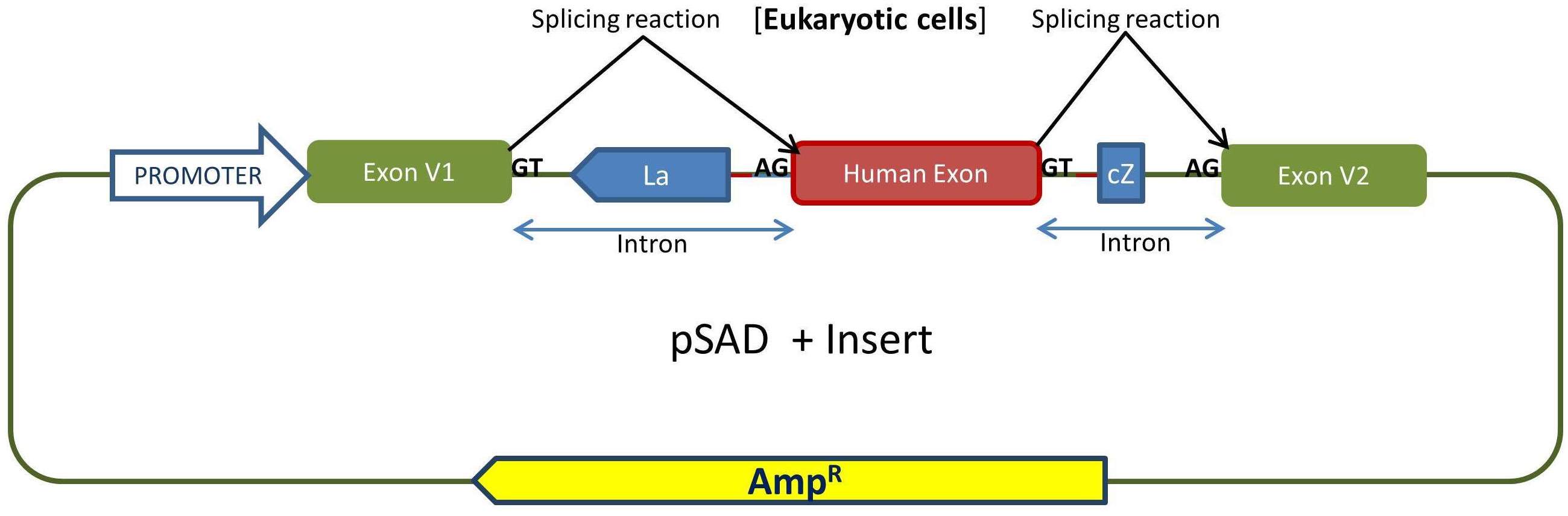
- Cloning of exon(s) of interest in pSAD. It is recommended to clone flanking exons to keep the genomic context that can influence on the splicing reactions. Alternatively, gene synthesis procedures can be carried out to circumvent this laborious step.
Outline of the cloning procedure into the pSAD vector. The insert with exon(s) is generated by PCR with a high fidelity polymerase and primers with tails containing suitable restriction sites to keep the orientation of acceptor and donor sites with those of the constitutive exons of pSAD (V1 and V2). E. coli DH5α cells are transformed and selected with ampicillin and X-Gal/IPTG. The splicing reactions in eukaryotic cells are indicated.
- Generation of the DNA variant by site directed-mutagenesis;
- Transfection of the wild type and mutant minigenes into eukaryotic cells;
- RNA Isolation and RT-PCR with specific primers of the constitutive exons (V1 and V2) that do not amplify RNA transcribed by the host cell;
- Electrophoresis and sequencing of RNA isoforms: characterization of anomalous events (e.g. exon skipping, use of alternative splice sites, intron retention, etc).
 The full protocol (cloning and functional test) would take 6-12 weeks depending on the number of the cloning steps. Once the wild type minigene has been constructed, it can serve as a template to introduce and test any candidate DNA variant by PCR-mutagenesis so the experimental time is significantly reduced.
The full protocol (cloning and functional test) would take 6-12 weeks depending on the number of the cloning steps. Once the wild type minigene has been constructed, it can serve as a template to introduce and test any candidate DNA variant by PCR-mutagenesis so the experimental time is significantly reduced.
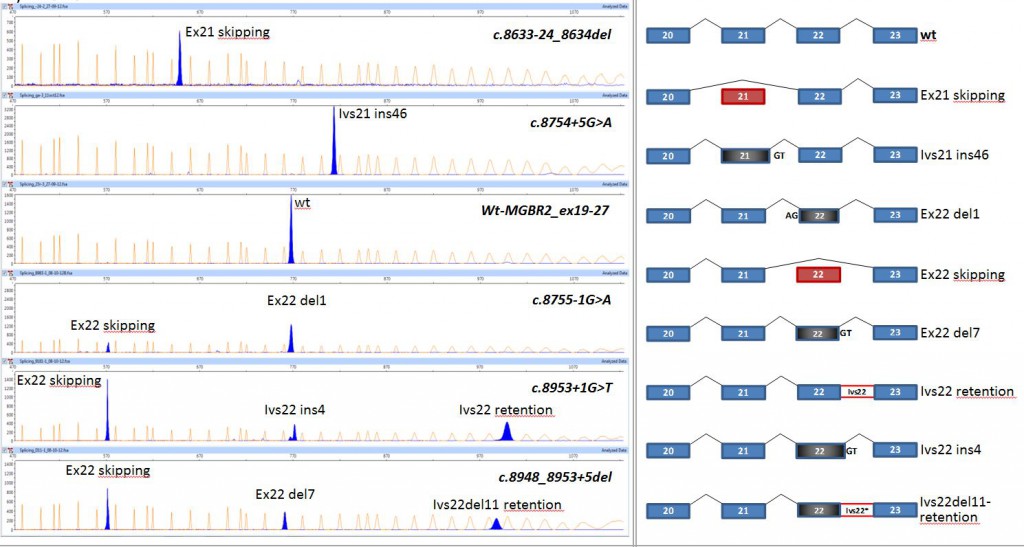
BRCA2 EXONS 21-22
Splicing functional assays of BRCA2 minigene MGBR2_ex19-27: Impact on splicing of DNA variants of the acceptor and donor sites of exons 21 and 22. A) Capillary electrophoresis of fluorescent RT-PCRs
List of constructed minigenes
Breast cancer minigenes:
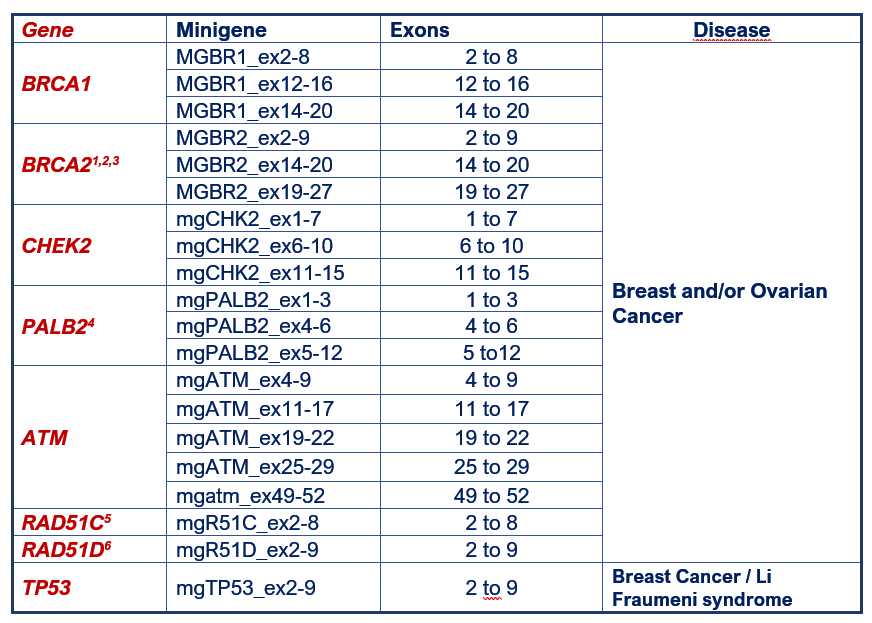
1 Fraile-Bethencourt et al (2019); 2 Fraile-Bethencourt et al (2017); 3 Acedo et al (2015); 4 Valenzuela-Palomo et al (2022); 5 Sanoguera-Miralles et al (2020); 6 Bueno-Martínez et al (2021).
The CHEK2, PALB2, ATM, RAD51C and RAD51D minigenes have been developed under the H2020 grant “Breast Cancer Risk after Diagnostic Gene Sequencing (BRIDGES)” (Project Number 634935).
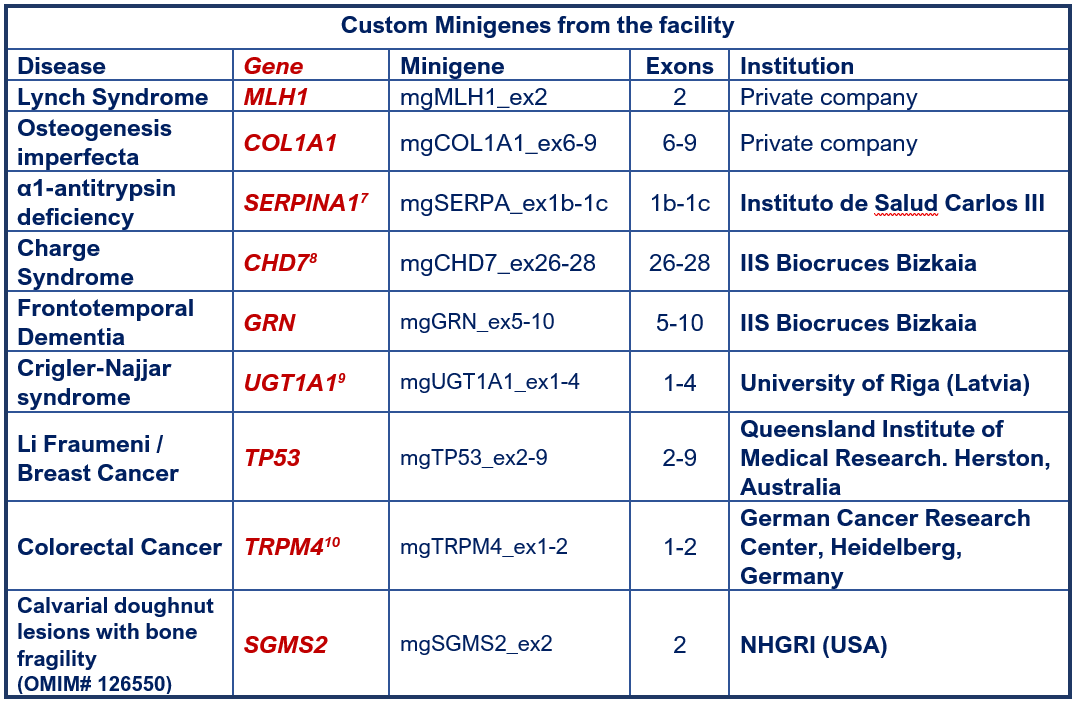
FACILITY MINIGENES
7 Lara et al (2014); 8 Villate et al (2018); 9 Gailite et al (2020); 10 Zhu et al (2022).


 Price List (24/05/2024)1
Price List (24/05/2024)1
- Bioinformatics analysis of DNA variants 50 €
- Analysis of patient samples:
- RNA extraction from blood 50 €
- Lymphocyte RT-PCR 300 €
- Splicing functional assays with hybrid minigenes:
- Construction of 1 custom minigene2 450 € (each cloning step, max. 3)
- Functional assay of a DNA variant 450 €
1 VAT not included.
2 Any human exon can be cloned.
Dr. Eladio A. Velasco-Sampedro (eavelsam@uva.es) should be contacted for any functional test.
REFERENCES- Acedo A, Sanz DJ, Duran M, Infante M, Perez-Cabornero L, Miner C, Velasco EA. 2012. Comprehensive splicing functional analysis of DNA variants of the BRCA2 gene by hybrid minigenes. Breast Cancer Res 14:R87. [PubMed]
- Acedo A, Hernández-Moro C, Curiel-García Á, Díez-Gómez B, Velasco EA. 2015. Functional Classification of BRCA2 DNA Variants by Splicing Assays in a Large Minigene with 9 Exons. Human Mutation 36: 210–221. [PubMed]
- Baralle D, Lucassen A, Buratti E. 2009. Missed threads. The impact of pre-mRNA splicing defects on clinical practice. EMBO Rep 10:810-816. [PubMed]
- Bueno-Martínez E, Sanoguera-Miralles L, Valenzuela-Palomo A, Lorca V, Gómez-Sanz A, Carvalho S, Allen J, Infante M, Pérez-Segura P, Lázaro C, Easton DF, Devilee P, Vreeswijk MPG, de la Hoya M, Velasco EA. 2021. RAD51D Aberrant Splicing in Breast Cancer: Identification of Splicing Regulatory Elements and Minigene-Based Evaluation of 53 DNA Variants. Cancers (Basel) 13:2845. doi: 10.3390/cancers13112845.
- Fraile-Bethencourt E, Díez-Gómez B, Velásquez-Zapata V, Acedo A, Sanz DJ, Velasco EA. 2017. Functional classification of DNA variants by hybrid minigenes: Identification of 30 spliceogenic variants of BRCA2 exons 17 and 18. PLoS Genetics 13(3):e1006691. doi: 10.1371/journal.pgen.1006691. http://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1006691
- Fraile-Bethencourt E, Valenzuela-Palomo A, Díez-Gómez B, Goina E, Acedo A, Buratti E, Velasco EA. 2019. Mis-splicing in breast cancer: identification of pathogenic BRCA2 variants by systematic minigene assays. J Pathol.; 248: 409-420. doi: 10.1002/path.5268. [Pubmed]
- Gailite L, Valenzuela-Palomo A, Sanoguera-Miralles L, Rots D, Kreile M and Velasco EA. UGT1A1 Variants c.864+5G>T and c.996+2_996+5del of a Crigler-Najjar Patient Induce Aberrant Splicing in Minigene Assays.Front. Genet. 2020; 11:169. doi: 10.3389/fgene.2020.00169. Front Genet 2020; 11: 169
- Lara B, Martínez MT, Blanco I, Hernández-Moro C, Velasco EA, Ferrarotti I, Rodriguez-Frias F, Perez L, Vazquez I, Alonso J, Posada M, Martínez-Delgado B. 2014. Severe alpha-1 antitrypsin deficiency in composite heterozygotes inheriting a new splicing mutation QOMadrid. Respiratory Research 15: 125. [PubMed]
- Lopez-Bigas N, Audit B, Ouzounis C, Parra G, Guigo R. 2005. Are splicing mutations the most frequent cause of hereditary disease? FEBS Lett 579:1900-1903. [PubMed]
- Ruiz de Garibay G, Acedo A, García-Casado Z, Gutiérrez-Enríquez S, Tosar A, Romero A, Garre P, Llort G, Thomassen M, Díez O, Pérez-Segura P, Eduardo Díaz-Rubio E, Velasco EA, Caldés T, de la Hoya M. 2014. Capillary Electrophoresis Analysis of Conventional Splicing Assays: IARC Analytical and Clinical Classification of 31 BRCA2 Genetic Variants. Human Mutation 35: 53-57. [PubMed]
- Sanoguera-Miralles L, Valenzuela-Palomo A, Bueno-Martínez E, Llovet P, Díez-Gómez B, Caloca MJ, Pérez-Segura P, Fraile-Bethencourt E, Colmena M, Carvalho S, Allen J, Easton DF, Devilee P, Vreeswijk MPG, de la Hoya M, Velasco EA. 2020. Comprehensive Functional Characterization and Clinical Interpretation of 20 Splice-Site Variants of the RAD51C Gene. Cancers (Basel) 12:3771. doi: 10.3390/cancers12123771.
- Sanz DJ, Acedo A, Infante M, Duran M, Perez-Cabornero L, Esteban-Cardenosa E, Lastra E, Pagani F, Miner C, Velasco EA. 2010. A high proportion of DNA variants of BRCA1 and BRCA2 is associated with aberrant splicing in breast/ovarian cancer patients. Clin Cancer Res 16:1957-1967. [PubMed]
- Valenzuela-Palomo A, Bueno-Martínez E, Sanoguera-Miralles L, Lorca V, Fraile-Bethencourt E, Esteban-Sánchez A, Gómez-Barrero S, Carvalho S, Allen J, García-Álvarez A, Pérez-Segura P, Dorling L, Easton DF, Devilee P, Vreeswijk MP, de la Hoya M, Velasco EA. 2022. Splicing predictions, minigene analyses, and ACMG-AMP clinical classification of 42 germline PALB2 splice-site variants. J Pathol. 256:321-334. doi: 10.1002/path.5839.
- Villate O; Ibarluzea N; Fraile-Bethencourt E; Valenzuela A; Velasco EA; Grozeva D; Raymond FL; Botella MP; Tejada MI. 2018. Functional analyses of a novel splice variant in the CHD7 gene, found by Next Generation Sequencing, confirm its pathogenicity in a Spanish patient and diagnose him with CHARGE syndrome. Frontiers in Genetics 2018, 9: 7. DOI: 10.3389/fgene.2018.00007
- Zhu L, Miao B, Dymerska D, Kuswik M, Bueno-Martínez E, Sanoguera-Miralles L, Velasco EA, Paramasivam N, Schlesner M, Kumar A, Yuan Y, Lubinski J, Bandapalli OR, Hemminki K, Försti A. 2022. Germline Variants of CYBA and TRPM4 Predispose to Familial Colorectal Cancer. Cancers (Basel) 14:670. doi: 10.3390/cancers14030670.